JNets 1.0
:: DESCRIPTION
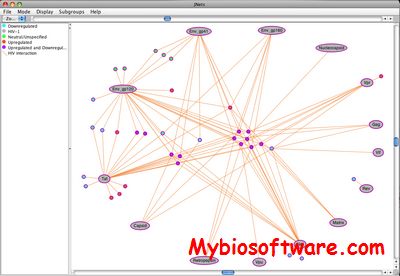
JNets is a network visualization and analysis tool. JNets was primarily designed for visualizing and analyzing protein interaction networks, but is flexible enough to be used for any type of network. JNets is available as a stand alone application or an applet. Using the applet, interactive networks can be deployed from web pages.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2009 Mar 26;10:95.
JNets: exploring networks by integrating annotation.
Macpherson JI, Pinney JW, Robertson DL.