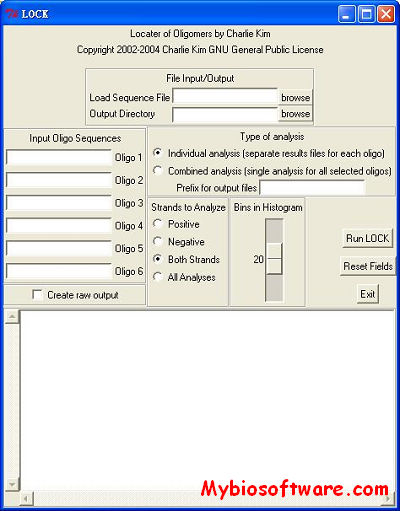
LOCK 2.0
:: DESCRIPTION
LOCK is a vector based protein structure superposition algorithm capable of recognizing distant structural similarities. Statistical significance values for alignments are provided.
::DEVELOPER
The Brutlag Bioinformatics Group
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- C Complier
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Amit P. Singh ,Douglas L. Brutlag
Hierarchical Protein Structure Superposition using both Secondary Structure and Atomic Representations
Proceedings of the 5th International Conference on Intelligent Systems for Molecular Biology