jMODELTEST 2.1.10
:: DESCRIPTION
jModelTest is a tool to carry out statistical selection of best-fit models of nucleotide substitution. It implements five different model selection strategies: hierarchical and dynamical likelihood ratio tests (hLRT and dLRT), Akaike and Bayesian information criteria (AIC and BIC), and a decision theory method (DT). It also provides estimates of model selection uncertainty, parameter importances and model-averaged parameter estimates, including model-averaged phylogenies. Please note that the DT weights are very gross.
jModelTest2 includes High Performance Computing (HPC) capabilities and additional features like new strategies for tree optimization, model-averaged phylogenetic trees (both topology and branch length), heuristic filtering and automatic logging of user activity.
::DEVELOPER
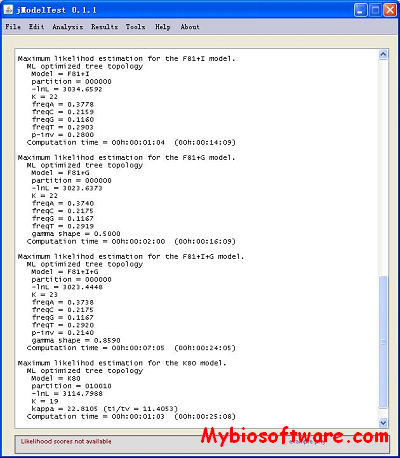
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOS
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Posada D. 2008.
jModelTest: Phylogenetic Model Averaging.
Molecular Biology and Evolution 25: 1253-1256.
Darriba D, Taboada GL, Doallo R, Posada D. 2012.
jModelTest 2: more models, new heuristics and parallel computing.
Nature Methods 9(8), 772.