SAFA 1.1
:: DESCRIPTION
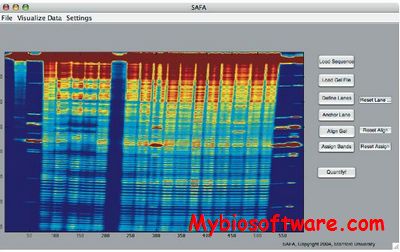
SAFA (Semi-Automated Footprinting Analysis) is a softwre for rapidly quantifying the band intensities from nucleic acid chemical mapping gels at single nucleotide resolution.The protocols implemented in SAFA have five steps: 1.) Lane identification, 2.) Gel rectification, 3.) Band assignment, 4.) Model fitting, and 5.) Band intensity normalization. SAFA enables the rapid quantitation of gel images containing thousands of discrete bands, thereby eliminating a bottleneck to the analysis of chemical mapping experiments. An experienced user of the software can quantify a gel image in approximately 15 minutes. Although SAFA was developed to analyze hydroxyl radical (·OH) footprints, it effectively quantifies the gel images obtained with other types of chemical mapping probes. We also present a series of tutorial movies that illustrate the best practices and different steps in the SAFA analysis as a supplement to this protocol.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/Windows/MacOsX
:: DOWNLOAD
:: MORE INFORMATION
Citation
Laederach, A., Das, R., Vicens, Q., Pearlman, S.M., Brenowitz, M., Herschlag, D., and Altman, R.B. (2008)
Semi-automated and rapid quantification of nucleic acid footprinting and structure mapping experiment.
Nature Protocols 3(9), 1395-1401. (2008)