EASE 2.0
:: DESCRIPTION
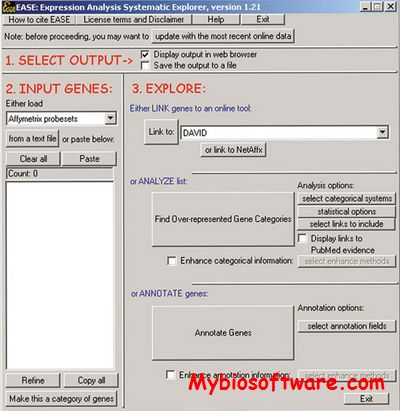
EASE(the Expression Analysis Systematic Explorer), developed by DAVID Bioinformatics team, is a customizable, standalone, Windows(c) desktop software application that facilitates the biological interpretation of gene lists derived from the results of microarray, proteomic, and SAGE experiments. EASE provides statistical methods for discovering enriched biological themes within gene lists, generates gene annotation tables, and enables automated linking to online analysis tools.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Douglas A. Hosack, Glynn Dennis Jr., Brad T. Sherman, H. Clifford Lane, Richard A. Lempicki.
Identifying Biological Themes within Lists of Genes with EASE.
Genome Biology 2003 4(6):P4.