Metingear 1.1.6
:: DESCRIPTION
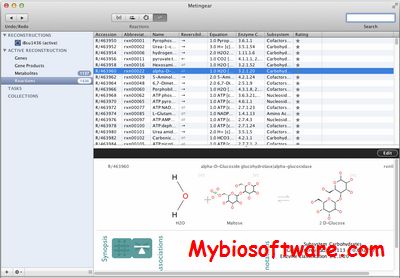
Metingear is an open source desktop application for creating and curating genome scale metabolic networks with chemical structure.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux/ MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2013 Sep 1;29(17):2213-5. doi: 10.1093/bioinformatics/btt342. Epub 2013 Jun 13.
Metingear: a development environment for annotating genome-scale metabolic models.
May JW1, James AG, Steinbeck C.