Gambit 0.4.145
:: DESCRIPTION
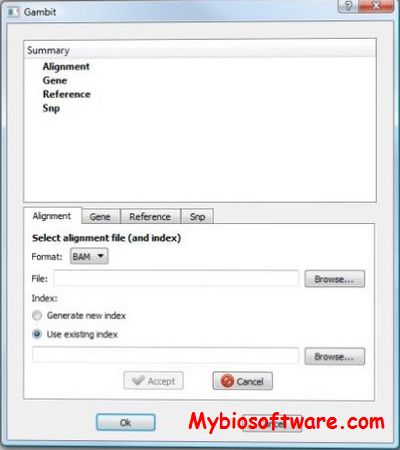
Gambit is a new cross-platform GUI (graphical user interface) application for sequence visualization and analysis.The software takes advantage of the indexing features of the (fairly) recently standardized BAM sequence alignment format that allows rapid access to genomic data, with minimal startup time and re-rendering delay. Gambit also supports a variety of annotation formats (BED, GFF, GFF3, VCF) “out-of-the-box” for displaying gene/region annotations as well as SNP entries.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/windows/MacOsX
:: DOWNLOAD
:: MORE INFORMATION