Melting 5.2.0
:: DESCRIPTION
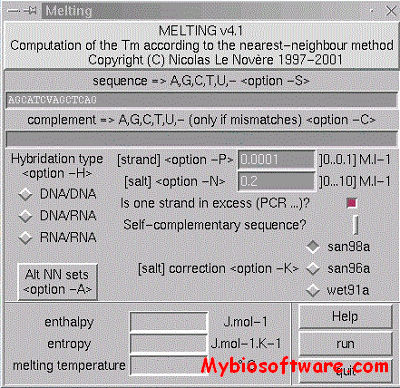
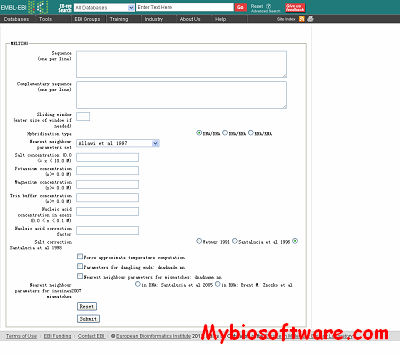
Melting computes, for a nucleic acid duplex, the enthalpy, the entropy and the melting temperature of the helix-coil transitions.Three types of hybridisation are possible: DNA/DNA, DNA/RNA, and RNA/RNA. The program first computes the hybridisation enthalpy and entropy from the elementary parameters of each Crick’s pair by the nearest-neighbor method. Then the melting temperature is computed. The set of thermodynamic parameters can be easely changed, for instance following an experimental breakthrough.
MELTING provides a large set of thermodynamic models to compute the enthalpy and entropy of several structures in the duplex : perfectly matching sequences, single mismatch, tandem mismatch, internal loop, single dangling end, second dangling end, long dangling end (only one to four poly A), single bulge loop, long bulge loop, inosine base (I), hydroxyadenine (A*), azobenzene (cis X_C and trans X_T), locked nucleic acids (Al, Gl, Tl, Cl).
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2012 May 16;13:101. doi: 10.1186/1471-2105-13-101.
MELTING, a flexible platform to predict the melting temperatures of nucleic acids.
Dumousseau M1, Rodriguez N, Juty N, Le Novère N.