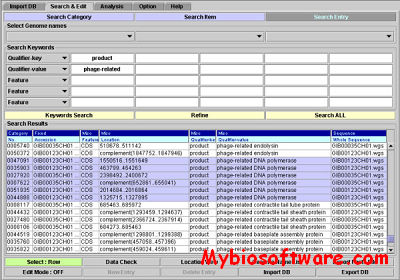
G-InforBIO 1.90
:: DESCRIPTION
G-InforBIO is an e-Workbench for “databasing”, comparative genome analysis. The G-InforBIO system is a novel tool for genome data management and sequence analysis. The system can import genome data encoded as eXtensible Markup Language documents as formatted text documents, including annotations and sequences, from DNA Data Bank of Japan and GenBank encoded as flat files. The genome database is constructed automatically after importing, and the database can be exported as documents formatted with eXtensible Markup Language or tab-deliminated text. Users can retrieve data from the database by keyword searches, edit annotation data of genes, and process data with G-InforBIO. In addition, information in the G-InforBIO database can be analyzed seamlessly with nine different software programs, including programs for clustering and homology analyses.
::DEVELOPER
WFCC-MIRCEN World Data Centre for Microorganisms
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / Mac OsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Naoto Tanaka, Takashi Abe, Satoru Miyazaki and Hideaki Sugawara,
“G-InforBIO: Integrated system for microbial genomics“,
BMC Bioinformatics, 7, 368, (2006).