DRACK 3.1
:: DESCRIPTION
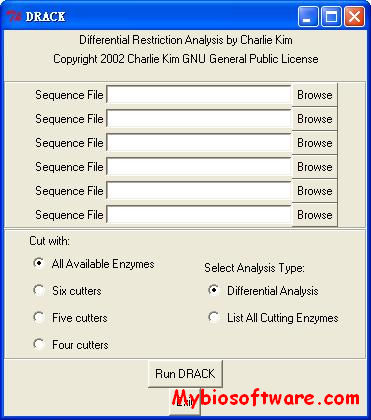
DRACK takes up to 6 FASTA DNA sequence files as input and outputs a tab-delimited text file containing sizes of restriction fragments (which can be opened in a spreadsheet program). Several options are available, including 4-6 base cutters and running in differential or list-all cutters mode. Differential analysis will analyze the sequences and only output restriction enzymes which distinguish between the sequences, while listing all cutters will list even those cutters which do not distinguish between the sequences. This program’s primary purpose was to automatically choose sites to distinguish plasmid clones with an insert in two possible orientations.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
The software is copyrighted under the terms of the GNU General Public License. You can view this license at http://www.gnu.org/licenses/gpl.txt.