Bioclipse 2.6.2
:: DESCRIPTION
Bioclipse is a free and open source workbench for the life sciences with a state-of-the-art plugin architecture, enabling powerful integration of existing software, and making it easy to customize for different needs.
Bioclipse has advanced functionality in fields such as cheminformatics, bioinformatics, semantic web, spectrum analysis, drug discovery, safety assessment and general chemistry education.
Features
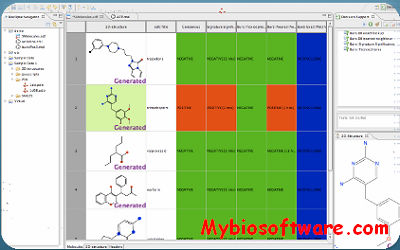
- The Bioclipse-QSAR feature makes descriptor calculation and formation of QSAR datasets easy and reproducible, adhering to the open QSAR-ML standard. Bioclipse is equipped with graphical editors and wizards, and supports a wide range of chemical descriptors.
- Bioclipse contains advanced functionality to import, edit, and save chemical structures in various formats. Single molecules can be edited in an advanced 2D-editor, and the Chemistry Development Kit provides calculation of a wide range of properties, as well as generation of 2D and 3D structure, various fingerprints, InChI, and SMILES.
- Bioclipse can process large collections of molecules, primarily SD-files, in the range of GB’s. Bioclipse not only allows for browsing such large libraries, but also has means to edit individual compounds and then continue browsing.
- Bioclipse can calculate various properties on SD-files without opening them in the GUI. Such batch-calculations are faster, and can be carried out in teh background while continuing with other tasks.
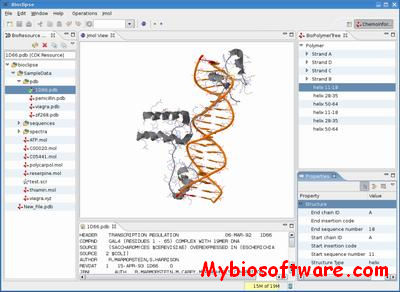
- Bioclipse has advanced, interactive 3D-visualization via the integrated Jmol application. Proteins and chemical compounds with 3D coordinates can be visualized with a multitude of rendering options.
- Predicting metabolic sites is important in the drug discovery process to aid in rapid compound optimization.
- The Bioclipse MetaPrint2D feature allows for rapid and accurate ranking of likely sites-of-metabolism, trained on phase-1 reactions in the Symyx Metabolite database. The predictions are very fast, less than 50 ms per molecule, and opens up for new ways of hypothesis testing in compound optimization. Calculations can be carried out on both individual molecules and on collections of compounds.
- Writing scripts in the Bioclipse Scripting Language (BSL) to create reusable functions that can be used to automate repetitive tasks and shared with others. BSL is based on JavaScript, and is very powerful in chemical and biological analyses. Bioclipse makes use of Gist and MyExperiment for sharing scripts.
- Easily download chemical structures from public repositories, such as PubChem, ChemSpider, and PDB.
- Use one of the available Wizards or include a snippet in your script to access chemical resources online.
::DEVELOPER
Bioclipse team
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
- Linux
- MacOsX
:: DOWNLOAD
:: MORE INFORMATION
If you use Bioclipse in your research, please cite:
Bioclipse: An open source workbench for chemo- and bioinformatics
Ola Spjuth, Tobias Helmus, Egon L Willighagen, Stefan Kuhn, Martin Eklund, Johannes Wagener, Peter Murray-Rust, Christoph Steinbeck, Jarl E.S. Wikberg
BMC Bioinformatics 2007, 8:59 doi:10.1186/1471-2105-8-59 Fulltext
and
Bioclipse 2: A scriptable integration platform for the life sciences
Ola Spjuth, Jonathan Alvarsson, Arvid Berg, Martin Eklund, Stefan Kuhn, Carl Mäsak, Gilleain Torrance, Johannes Wagener, Egon L Willighagen, Christoph Steinbeck and Jarl ES Wikberg
BMC Bioinformatics 2009, 10:397 doi:10.1186/1471-2105-10-397 Fulltext