TreeJuxtaposer 2.1
:: DESCRIPTION
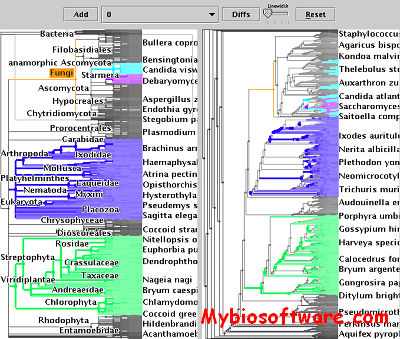
TreeJuxtaposer lets you interactively view a single tree, or compare two or more trees. The software handles all kinds of trees, including phylogenetic trees, taxonomic trees, consensus trees, dendrograms from cluster hierarchies, and others. Uses nested parentheses (Newick/New Hampshire) data format. Guaranteed visibility of marked regions provides visual landmarks. Structural differences are automatically marked, and the marked clades highlight corresponding nodes on the trees through the use of a generalization of the Robinson-Foulds distance metric. Responsive drawing for even very large trees with advanced graphics techniques.
::DEVELOPER
TreeJuxtaposer Team
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOSX
- Java
:: DOWNLOAD
 TreeJuxtaposer
TreeJuxtaposer
:: MORE INFORMATION
Citation:
Tamara Munzner, Francois Guimbretiere, Serdar Tasiran, Li Zhang, and Yunhong Zhou.
TreeJuxtaposer: Scalable Tree Comparison using Focus+Context with Guaranteed Visibility
Proc. SIGGRAPH 2003, published as ACM Transactions on Graphics 22(3), pages 453–462