Fastbreak
:: DESCRIPTION
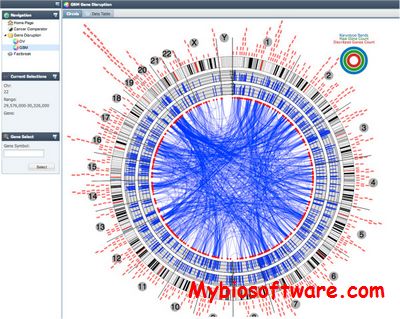
Fastbreak is a set of python scripts implementing an O(n) algorithm for detecting translocations and other genetic structural variants from aligned mate pair sequence data in SAM/BAM format.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Web Server
- Tomcat
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
EURASIP J Bioinform Syst Biol. 2012 Oct 9;2012(1):15. doi: 10.1186/1687-4153-2012-15.
Fastbreak: a tool for analysis and visualization of structural variations in genomic data.
Bressler R, Lin J, Eakin A, Robinson T, Kreisberg R, Rovira H, Knijnenburg T, Boyle J, Shmulevich I.