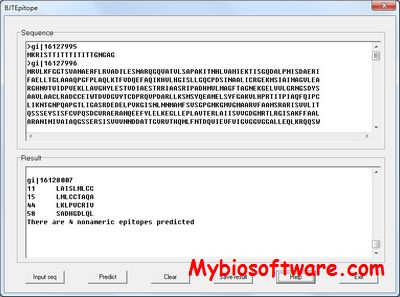
EpiPred
:: DESCRIPTION
EpiPred is a software of B-cell epitope prediction and antibody-antigen docking
::DEVELOPER
Oxford Protein Informatics Group (OPIG)
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows/ MacOsX
- JRE
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Improving B-cell epitope prediction and its application to global antibody-antigen docking.
Krawczyk K, Liu X, Baker T, Shi J, Deane CM.
Bioinformatics. 2014 Aug 15;30(16):2288-94. doi: 10.1093/bioinformatics/btu190.