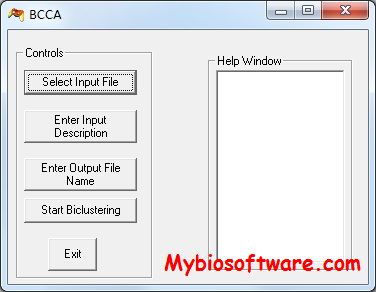
ClusterEnG
:: DESCRIPTION
ClusterEnG (acronym for Clustering Engine for Genomics) is an educational web resource on clustering and visualization of high-dimensional datasets. The resource currently offers visualization of PCA, t-SNE vectors of input dataset for several clustering algorithms. Furthermore, the user can also explore eighteen internal clustering validation measures to compare different clustering results.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web browser
:: DOWNLOAD
:: MORE INFORMATION
Citation
Manjunath M, Zhang Y, Yeo SH, Sobh O, Russell N, Followell C, Bushell C, Ravaioli U, Song JS.
ClusterEnG: an interactive educational web resource for clustering and visualizing high-dimensional data.
PeerJ Comput Sci. 2018;4:e155. doi: 10.7717/peerj-cs.155. Epub 2018 May 21. PMID: 30906871; PMCID: PMC6429934.