Bi-Force v2
:: DESCRIPTION
Bi-Force is a novel way of modeling the problem as combinatorial optimization problem on graphs: Weighted Bi-Cluster Editing. It is a very flexible model that can handle arbitrary kinds of multi-condition data sets (not limited to gene expression).
::DEVELOPER
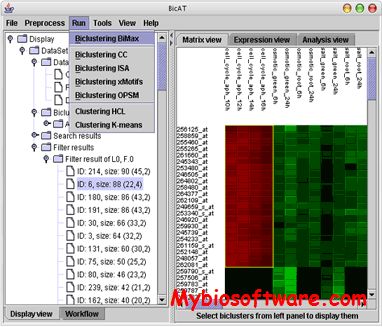
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bi-Force: large-scale bicluster editing and its application to gene expression data biclustering.
Sun P, Speicher NK, Röttger R, Guo J, Baumbach J.
Nucleic Acids Res. 2014 May;42(9):e78. doi: 10.1093/nar/gku201.