DECIPHER 2.20.0
:: DESCRIPTION
DECIPHER (Database Enabled Code for Ideal Probe Hybridization Employing R) is a software toolset that can be used for deciphering and managing DNA sequences efficiently using the R statistical programming language. The program is designed to be used with non-destructive workflows that guide the user through the process of importing, maintaining, analyzing, manipulating, and exporting a massive amount of DNA sequences.
::DEVELOPER
:: SCREENSHOTS
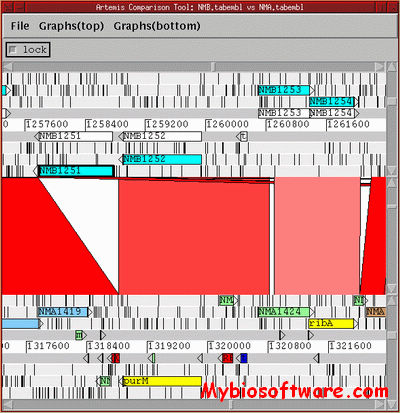
N/A
:: REQUIREMENTS
- Linux / Windows / MacOsX
- R package
- BioConductor
:: DOWNLOAD
:: MORE INFORMATION
Citation
DesignSignatures: a tool for designing primers that yield amplicons with distinct signatures.
Wright ES, Vetsigian KH.
Bioinformatics. 2016 Jan 23. pii: btw047
ES Wright et al. (2011)
“DECIPHER: A Search-Based Approach to Chimera Identification for 16S rRNA Sequences.”
Applied and Environmental Microbiology,