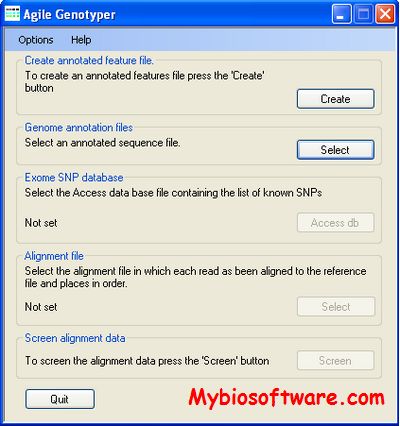
RepeatSeq v0.8.2
:: DESCRIPTION
RepeatSeq determines genotypes for microsatellite repeats in high-throughput sequencing data.
:: DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux/ Windows
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation
Nucleic Acids Res. 2013 Jan 7;41(1):e32. doi: 10.1093/nar/gks981. Epub 2012 Oct 22.
Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles.
Highnam G1, Franck C, Martin A, Stephens C, Puthige A, Mittelman D.