GMATA v2.3
:: DESCRIPTION
GMATA is an easiest and fastest bioinformatic tool /software for any Simple Sequence Repeats (SSR) analyses, and SSR marker designing, polymorphism screen, and e-mapping in any DNA sequences.
::DEVELOPER
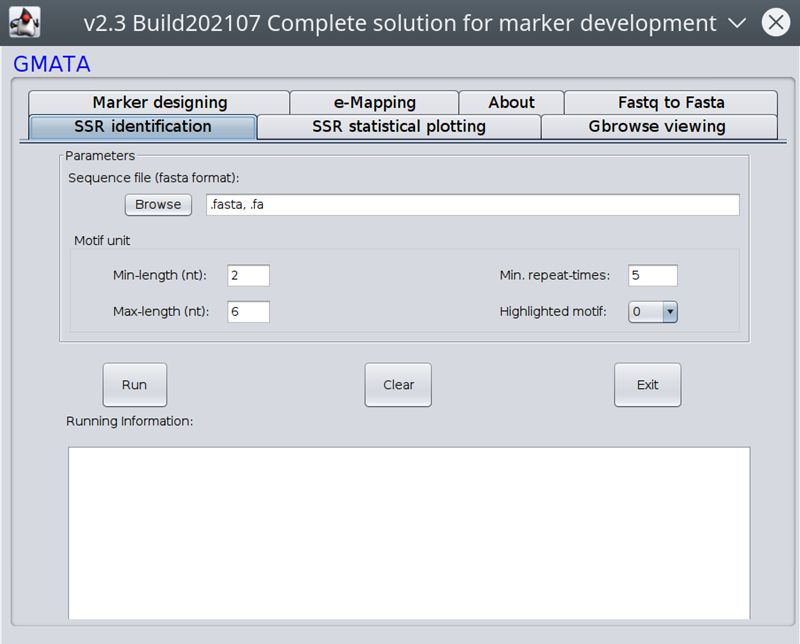
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
- Perl
:: DOWNLOAD
:: MORE INFORMATION
Citation
Wang X, Wang L.
GMATA: An Integrated Software Package for Genome-Scale SSR Mining, Marker Development and Viewing.
Front Plant Sci. 2016 Sep 13;7:1350. doi: 10.3389/fpls.2016.01350. PMID: 27679641; PMCID: PMC5020087.