JaDis
:: DESCRIPTION
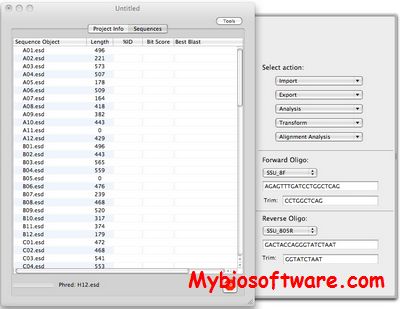
JaDis is a program for computing distances between nucleic acid sequences. It allows specific comparison of coding sequences, of non-coding sequences, or of coding and non-coding sequences.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/MacOsx/Linux
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Goncalves I, Robinson M, Perriere G, Mouchiroud D.
JaDis: computing distances between nucleic acid sequences.
Bioinformatics. 1999 May;15(5):424-5.