ePMV 0.5
:: DESCRIPTION
ePMV (embedded Python Molecular Viewer) runs molecular modeling software directly inside of professional 3D animation applications (hosts) to provide simultaneous access the capabilities of all of the systems. Uniting host and scientific algorithms into a single interface allows users from varied backgrounds to assemble professional quality visuals and to perform computational experiments with relative ease.
::DEVELOPER
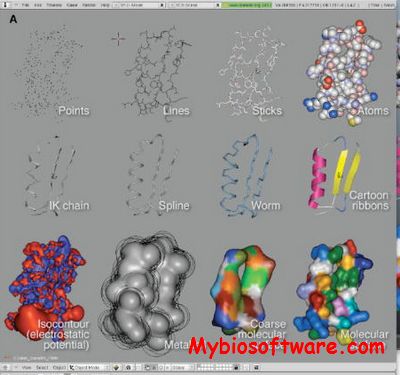
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / Mac OsX
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Johnson, G.T. and Autin, L., Goodsell, D.S., Sanner, M.F., Olson, A.J. (2011).
ePMV Embeds Molecular Modeling into Professional Animation Software Environments.
Structure 19, 293-303.