ChiLay 2.0
:: DESCRIPTION
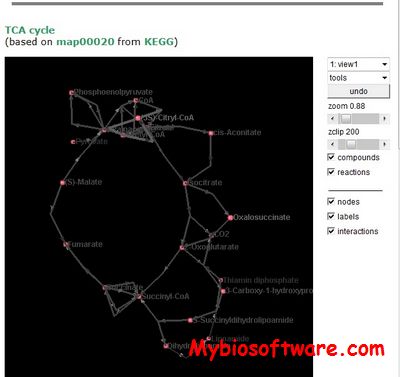
ChiLay (Chisio Layout) is a free, open source graphics/rendering independent Java component, which can be used by interactive graph editing tools to automatically layout compound or clustered graphs as well as simple, flat graphs
::DEVELOPER
i-Vis (Information Visualization) Research Group
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
An Algorithm for Automated Layout of Process Description Maps Drawn in SBGN.
Genc B, Dogrusoz U.
Bioinformatics. 2015 Sep 10. pii: btv516.