J3DPSV 1.0
:: DESCRIPTION
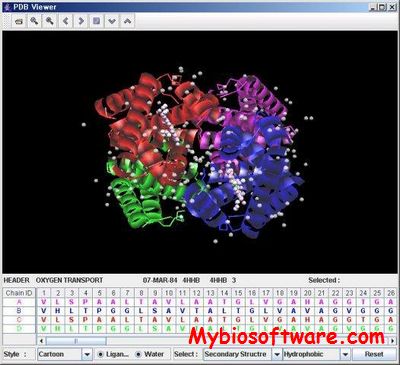
J3dPSV is a graphical application package for viewing and modeling of three dimensional structures of protein structure, including multiple chain sequence table and a three-dimensional (3D) protein structure viewer.
:: DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux / MacOsX
- JAVA
:: DOWNLOAD
:: MORE INFORMATION
Citation
Chun, Yoo Jin, Il Ham, Seong, Yang, San Duk, Rhie, Arang, Park, Hyun Seok
Refactoring the code for visualizing protein database information in a 3D viewer for software reusability
Genomics & Informatics, 6:50-53, MAR 31 2008