NeoBio 1.0 pre-alpha
:: DESCRIPTION
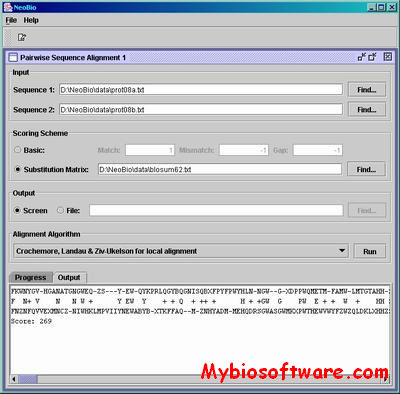
NeoBio is a Java class library of Computational Biology Algorithms. The current version consists mainly of pairwise sequence alignment algorithms such as the classical dynamic programming methods of Needleman-Wunsch and Smith-Waterman.
Advertisement
::DEVELOPER
Sérgio Anibal de Carvalho Junior
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux / Mac OsX
- JAVA
:: DOWNLOAD
:: MORE INFORMATION
NeoBio is free software, so feel free to download and use it at your own will.