SLIMS 1.0.0
:: DESCRIPTION
SLIMS is a powerful and user-friendly open source web application that provides all members of a laboratory with an interface to view, edit, and create sample information. SLIMS aims to simplify common laboratory tasks with tools such as a user-friendly shopping cart for subjects, samples and containers that easily generates reports, shareable lists, and plate designs for genotyping.
::DEVELOPER
:: SCREENSHOTS
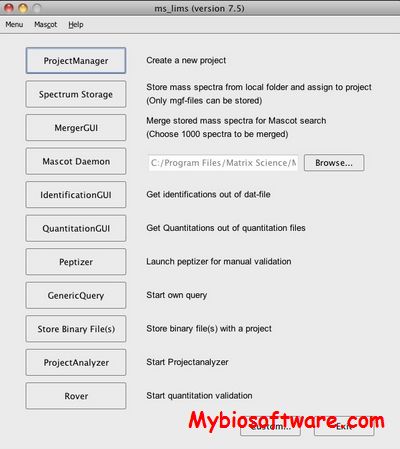
N/A
:: REQUIREMENTS
- Linux
- Apache
- MySQL
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Bioinformatics. 2010 Jul 15;26(14):1808-10. doi: 10.1093/bioinformatics/btq271. Epub 2010 May 30.
SLIMS–a user-friendly sample operations and inventory management system for genotyping labs.
Van Rossum T1, Tripp B, Daley D.