Evoker 2.2
:: DESCRIPTION
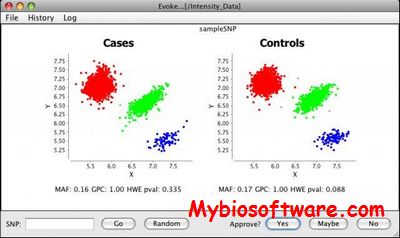
Evoker is a graphical tool for visualizing genotype intensity data in order to assess genotype calls as part of quality control procedures for genome-wide association studies.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Evoker: a visualization tool for genotype intensity data.
Morris JA, Randall JC, Maller JB and Barrett JC
Bioinformatics (Oxford, England) 2010;26;14;1786-7