ENT 1.0.2
:: DESCRIPTION
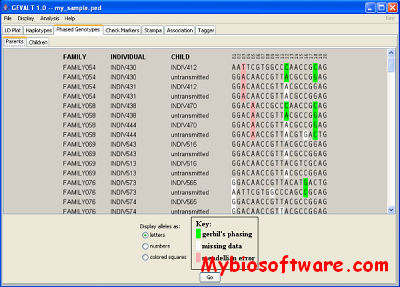
ENT is a highly scalable genotype phasing algorithm based on entropy minimization. ENT is capable of phasing both unrelated and related genotypes coming from complex pedigrees.
::DEVELOPER
Bioinformatics Lab , Computer Science & Engineering Dept. University of Connecticut
:: SCREENSHOTS
Command Line
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation:
A. Gusev and I.I. Mandoiu and B. Pasaniuc,
Highly Scalable Genotype Phasing by Entropy Minimization,
IEEE/ACM Trans. on Computational Biology and Bioinformatics 5, pp. 252-261, 2008