SGCaller
:: DESCRIPTION
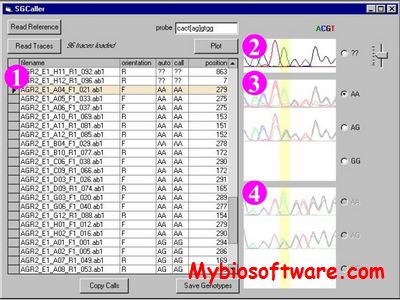
SGCaller is a Windows program that calls genotypes in sequencing traces.The software reads a collection of trace files in ABI or Staden (SCF) format, finds the variant position and tries to call the genotype for each, and presents them for the user to confirm and enhance the calling. SGCaller determines each file’s read direction from its name. After aligning sample traces to the reference, SGCaller calculates the total area under all four peaks for the base to be genotyped. Peak boundaries from the trace file are used; traces are not normalized to one another.
::DEVELOPER
Institute for Clinical Molecular Biology
:: SCREENSHOTS
::REQUIREMENTS
- Windows
- Microsoft Universal Data Access (MDAC)
:: DOWNLOAD
:: MORE INFORMATION
Citation
Manaster C, Valentonyte R, Teuber M, Zheng W, Schreiber S, Hampe J (2005).
SGCaller: A program to call and review genotypes measured by sequencing.
BioTechniques 38(4): 544-546