GenCo 200806
:: DESCRIPTION
GenCo is program to estimate gene conversion and crossover rates in population genetic data
::DEVELOPER
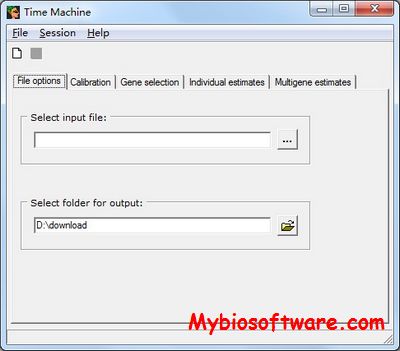
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Gay J, Myers S, and McVean G (2007).
Estimating meiotic gene conversion rates from population genetic data.
Genetics 177:881-94.