Miropeats 2.02
:: DESCRIPTION
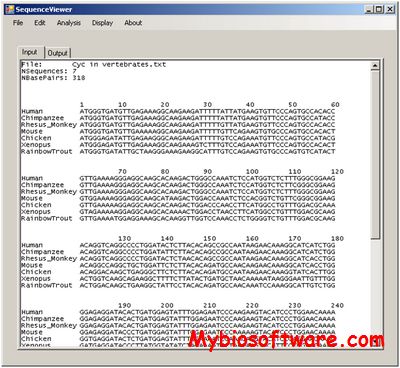
Miropeats discovers regions of sequence similarity amongst any set of DNA sequences and then presents this similarity information graphically. Sequence similarity searching is a very general tool that forms the basis of many different biological sequence analyses but it is limited by the verbosity of traditional alignment presentation styles. Miropeats enhances the utility of conventional DNA sequence comparisons when looking at long lengths of sequence similarity by summarizing extensive large scale sequence similarities on a single page of graphics.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- C Complier
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Comput Appl Biosci. 1995 Dec;11(6):615-9.
Miropeats: graphical DNA sequence comparisons.
Parsons JD.