GEB
:: DESCRIPTION
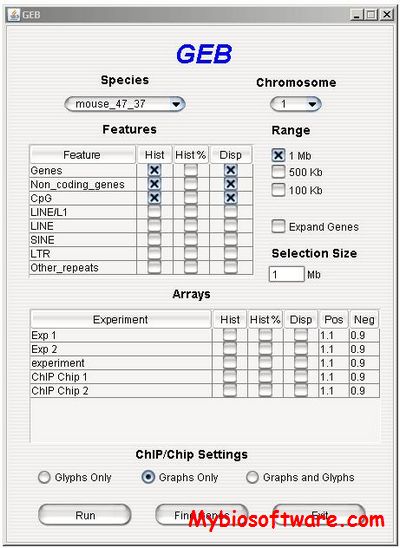
GEB is a Java application developed to provide a dynamic graphical interface to visualise the distribution of genome features and chromosome-wide experimental data in high resolution.
::DEVELOPER
the Bioinformatics Support Service, Centre for Bioinformatics, Imperial College London, UK and the MRC Clinical Sciences Centre, London, UK
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/ Windows/MacOsX
- Java
- MySQL
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2008 Nov 27;9:501. doi: 10.1186/1471-2105-9-501.
Genome Environment Browser (GEB): a dynamic browser for visualising high-throughput experimental data in the context of genome features.
Huntley D, Tang YA, Nesterova TB, Butcher S, Brockdorff N.