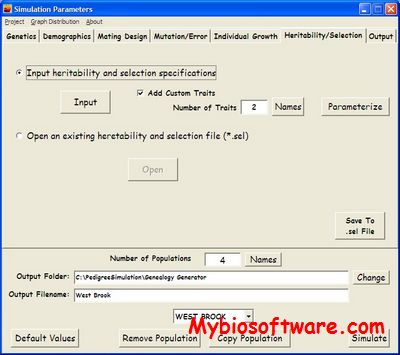
ExonScan 1.0
:: DESCRIPTION
ExonScan expects a primary transcript sequence, preferably excluding the first and last exon. It also currently needs at least about 20 bases upstream of the first exon it can locate, and at least about 60 bases downstream of the last. Numbers in the sequence are discarded, so you can cut and paste from, e.g., a GenBank file.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
:: MORE INFORMATION
Citation
Cell. 2004 Dec 17;119(6):831-45.
Systematic identification and analysis of exonic splicing silencers.
Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB.