ETAscape
:: DESCRIPTION
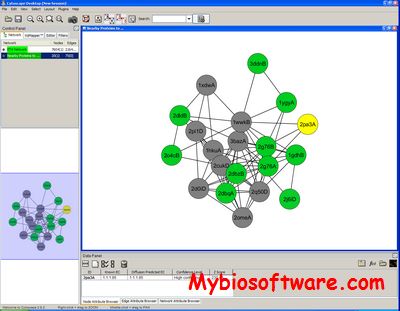
The ETAscape Cytoscape plugin allows users to make predictions and visualize the basis for predictions of protein function based on evolutionarily important structural motifs.
::DEVELOPER
Lichtarge Computational Biology Lab
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / Mac OsX
- Java
- Cytoscape
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Bioinformatics. 2012 Aug 15;28(16):2186-8. doi: 10.1093/bioinformatics/bts331. Epub 2012 Jun 11.
ETAscape: analyzing protein networks to predict enzymatic function and substrates in Cytoscape.
Bachman BJ, Venner E, Lua RC, Erdin S, Lichtarge O.