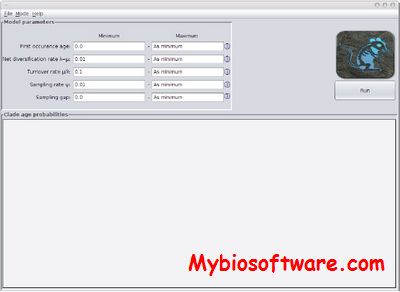
RMfam 0.3
:: DESCRIPTION
RMfam is a database of annotated multiple sequence alignments and probabilistic models for a number of RNA motifs.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- Perl
:: DOWNLOAD
:: MORE INFORMATION
Citation
Annotating RNA motifs in sequences and alignments.
Gardner PP, Eldai H.
Nucleic Acids Res. 2015 Jan;43(2):691-8. doi: 10.1093/nar/gku1327