FitchAln 20100321
:: DESCRIPTION
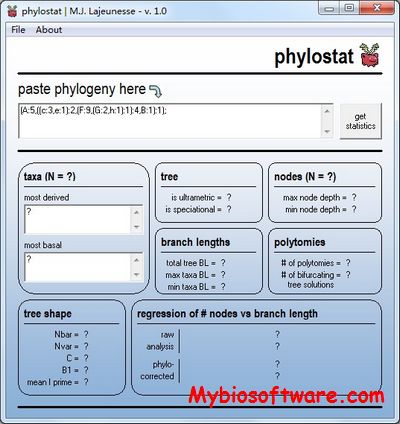
Given a newick binary tree T and its correpsonding multiple sequence alignemnt A of size n x m, where n is the number of leaf nodes in T and m is the length of the alignment, FitchAln generates a (n-1) x (m+1) Fitch score matrix representing the (Maximum Parsimony) number of mutations for each site for each internal node.
::DEVELOPER
UCF Computational Biology and Bioinformatics Group
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- C Compiler
:: DOWNLOAD
:: MORE INFORMATION