ViSEN 1.0beta0.3
:: DESCRIPTION
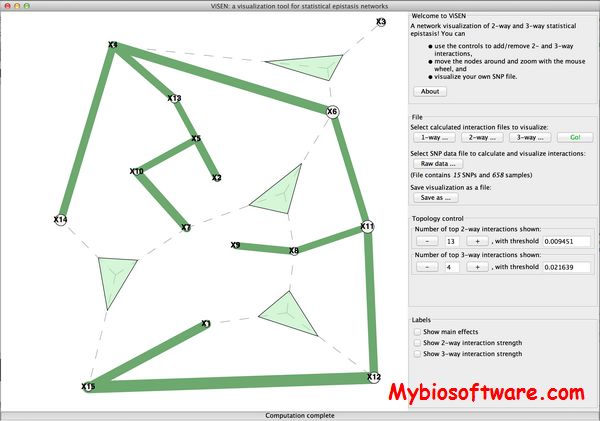
ViSEN provides a graphical visualization for statistical epistasis. Pairwise and three-way epistatic interactions are measured using information gain and are represented using networks.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Genet Epidemiol. 2013 Apr;37(3):283-5. doi: 10.1002/gepi.21718. Epub 2013 Mar 6.
ViSEN: methodology and software for visualization of statistical epistasis networks.
Hu T, Chen Y, Kiralis JW, Moore JH.