MAGNA++20150130
:: DESCRIPTION
MAGNA++ is a software tool for pairwise global network alignment. It is an extension of MAGNA. An example of an application domain where MAGNA++ is useful is computational biology – MAGNA++ can be used for biological network alignment.
MAGNA is an network alignment algorithm. MAGNA is a pairwise global aligner and a genetic algorithm. It can either start computation from precomputed alignments,or start completely from scratch.
::DEVELOPER
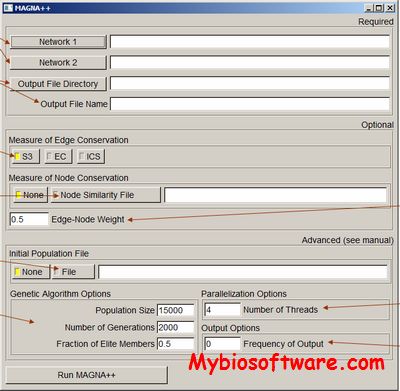
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / MacOsX
:: DOWNLOAD
:: MORE INFORMATION
Citation:
MAGNA++: Maximizing Accuracy in Global Network Alignment via both node and edge conservation.
Vijayan V, Saraph V, Milenković T.
Bioinformatics. 2015 Mar 19. pii: btv161
MAGNA: Maximizing Accuracy in Global Network Alignment.
Saraph V, Milenkovic T.
Bioinformatics. 2014 Jul 10. pii: btu409.