Arcadia 1.0rc2
:: DESCRIPTION
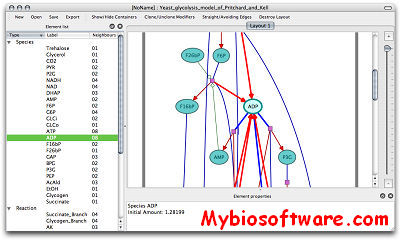
Arcadia is a light-weight, cross-platform, C++ desktop application designed for visualizing biological networks such as metabolic pathways.Arcadia is a viewer, not an editor: this means a simpler interface, offering multiple perspectives on the same data, with a focus on navigation. Arcadia translates text-based descriptions of biological networks (SBML files) into standardized diagrams (SBGN PD maps). Users can view the same model from different perspectives and easily alter the layout to emulate traditional textbook representations.
::DEVELOPER
Alice Villéger1,2, Steve Pettifer2, Douglas Kell1
[1] School of Chemistry, [2] School of Computer Science
at the University of Manchester, United Kingdom
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Mac OS X / Windows
- C ++ Complier
:: DOWNLOAD
:: MORE INFORMATION
Citation
Alice C. Villéger, Stephen R. Pettifer, Douglas B. Kell (2010)
Arcadia: a visualization tool for metabolic pathways,
Bioinformatics 2010 26(11):1470-1471; doi:10.1093/bioinformatics/btq154