silcLOD
:: DESCRIPTION
silcLOD (significance levels and critical LODs) is designed to calculate nominal significance levels and critical LOD scores depending on the length of the investigated region, number of chromosomes, and the cross-over rate. The global significance level as well as the precision of the calculation have to be specified.
::DEVELOPER
Institute of Medical Biometry and Statistics, University of Lübeck
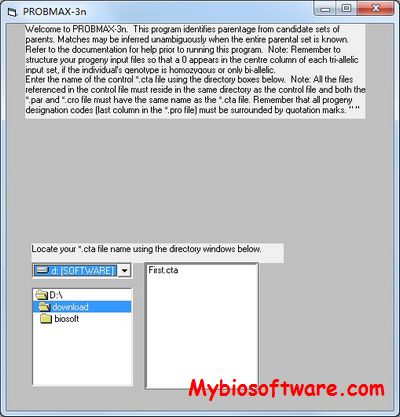
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
E Lander, L Kruglyak (1995),
“Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results”,
Nature Genetics, 11:241-247.