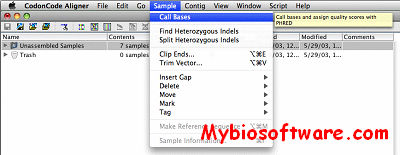
Admixem v1.0.2.2
:: DESCRIPTION
Admix’em (Admixture simulator) is a new forward-time simulator that allows for rapid and realistic simulations of admixed populations with selection.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / MacOsX
- C++ COmpiler
:: DOWNLOAD
:: MORE INFORMATION
Citation
Admix’em: A flexible framework for forward-time simulations of hybrid populations with selection and mate choice.
Cui R, Schumer M, Rosenthal GG.
Bioinformatics. 2015 Nov 28. pii: btv700.