VAGUE 1.0.5
:: DESCRIPTION
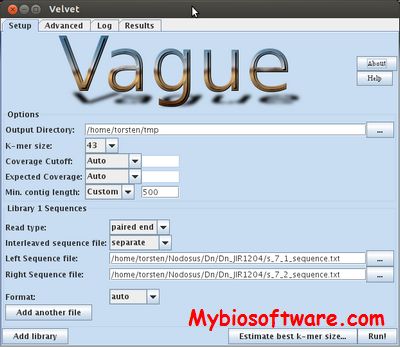
VAGUE (Velvet Assembler Graphical Front End) is a GUI for the Velvet de novo assembler.
::DEVELOPER
Victorian Bioinformatics Consortium
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX
- Java
- Velvet
:: DOWNLOAD
:: MORE INFORMATION
Citation
Powell DR, Seemann T.
VAGUE: a graphical user interface for the Velvet assembler.
Bioinformatics. 2013 Jan 15;29(2):264-5. doi: 10.1093/bioinformatics/bts664. Epub 2012 Nov 17. PMID: 23162059.