TREECON 1.3b
:: DESCRIPTION
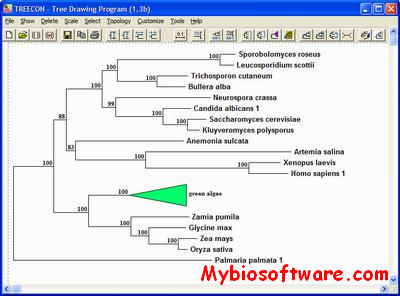
TREECON is a software package developed primarily for the construction and drawing of phylogenetic trees on the basis of evolutionary distances inferred from nucleic and amino acid sequences. The evolutionary distance is computed for all pairs of organisms (or sequences) and a phylogenetic tree is inferred by considering the relationship between these distance values. In pairwise distance methods, the dissimilarity (fraction of substitutions) is usually converted into evolutionary distance by correcting for multiple mutations. The most frequently used equations to convert dissimilarities into distances are implemented in TREECON. On the basis of these distances, a phylogenetic tree can be inferred. Different algorithms are available to construct a phylogenetic tree and a few of them are implemented in TREECON. Programs for rooting the unrooted evolutionary trees, for drawing the tree on the screen, and for saving the tree are also included, as well as several other tools. Starting from a simple ASCII text file, containing nucleic acid or amino acid sequences with gaps required for mutual alignment, one can produce publishable trees in a user-friendly and straightforward way.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Van de Peer, Y., De Wachter, Y. (1994)
TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment.
Comput. Applic. Biosci. 10, 569-70.