ncRNAClassifier 1.2
:: DESCRIPTION
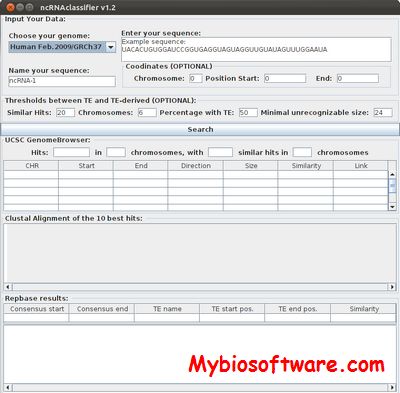
ncRNAClassifier is a tool enabling the identification of repetitive elements in precursor ncRNA sequences.
::DEVELOPER
EVRY RNA – IBISC
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2012 Sep 25;13:246. doi: 10.1186/1471-2105-13-246.
ncRNAclassifier: a tool for detection and classification of transposable element sequences in RNA hairpins.
Tempel S1, Pollet N, Tahi F.