Taxy / Taxy-Pro
:: DESCRIPTION
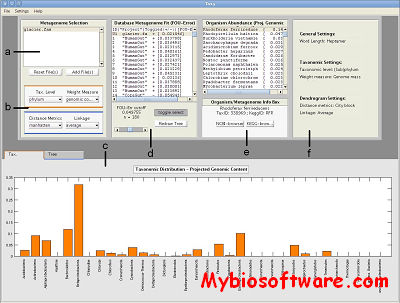
Taxy is a software for taxonomic profiling based on mixture modeling of the overall oligonucleotide distribution of a sample. Inferring the taxonomic composition of a microbial community from a large collection of anonymous DNA sequencing reads is a challenging task in computational biology. Because existing methods for taxonomic profiling of metagenomes are all based on the assignment of fragmental sequences to phylogenetic categories, the accuracy of results largely depends on fragment length. This dependency complicates comparative analysis of data originating from different sequencing platforms or preprocessing pipelines. We have developed a read length-independent method for taxonomic profiling and we provide a freely available Matlab/Octave toolbox which includes an ultra-fast implementation of that method. Besides the platform-independent toolbox we also provide a prototype tool implementation for Windows that allows the user to compare a large number of preprocessed metagenomes within a graphical environment.Our tests indicate that Taxy results compare well with taxonomic profiles obtained with other methods. However, in contrast to the existing methods, Taxy provides a nearly constant profiling accuracy across all kinds of read lengths and it operates at an unrivaled speed. As input, DNA sequences in terms of multi-FASTA files of any size can be used for the estimation of metagenomic profiles. The analysis of a large sequence file with a Gbp volume typically requires less than a minute of processing time and can even be performed on a standard notebook.
In contrast to the oligonucleotide-based Taxy method, Taxy-Pro is based on mixture model analysis of protein signatures in terms of protein domain frequencies.
::DEVELOPER
the Department of Bioinformatics of the University of Göttingen
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Mac OS X / Windows
- Matlab
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2013 Apr 15;29(8):973-80. doi: 10.1093/bioinformatics/btt077. Epub 2013 Feb 15.
Protein signature-based estimation of metagenomic abundances including all domains of life and viruses.
Klingenberg H1, Aßhauer KP, Lingner T, Meinicke P.
P. Meinicke, K. Asshauer and T. Lingner.
“Mixture models for analysis of the taxonomic composition of metagenomes“,
Bioinformatics May 15, 2011 27 (10)