Skyline 21.1
:: DESCRIPTION
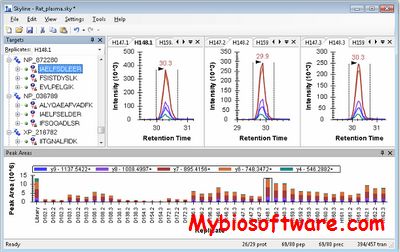
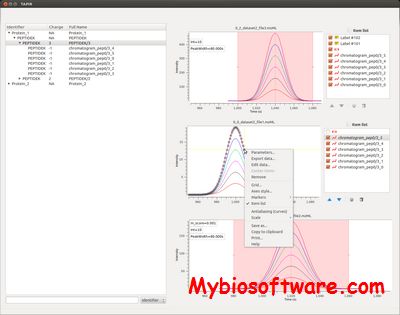
Skyline is a freely-available, open-source Windows client application for building Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM), Parallel Reaction Monitoring (PRM – Targeted MS/MS and DIA/SWATH) and targeted DDA with MS1 quantitative methods and analyzing the resulting mass spectrometer data. It aims to employ cutting-edge technologies for creating and iteratively refining targeted methods for large-scale proteomics studies.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2014 May 9. [Epub ahead of print]
A framework for installable external tools in Skyline.
Broudy D1, Killeen T, Choi M, Shulman N, Mani DR, Abbatiello SE, Mani D, Ahmad R, Sahu AK, Schilling B, Tamura K, Boss Y, Sharma V, Gibson BW, Carr SA, Vitek O, Maccoss MJ, Maclean B.