NET-SYNTHESIS
:: DESCRIPTION
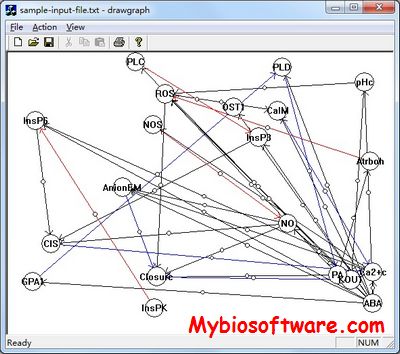
NET-SYNTHESIS performs combined synthesis, inference and simplification of signal transduction networks. The main idea of the application lies in representing observed indirect causal relationships as network paths, introducing pseudo-vertices for unknown intermediaries of these paths and using techniques from combinatorial optimization to find the most parsimonious graph consistent with all experimental observations. The software contains algorithms for (i) transitive reduction of an initially synthesized graph subject to the constraints that no edges corresponding to direct interactions are eliminated and (ii) pseudo-vertex collapse subject to the constraints that real (known) vertices are not eliminated.
::DEVELOPER
Bhaskar DasGupta
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
 NET-SYNTHESIS
NET-SYNTHESIS
:: MORE INFORMATION
Citation
Sema Kachalo, Ranran Zhang, Eduardo Sontag, Réka Albert and Bhaskar DasGupta,
NET-SYNTHESIS: A software for synthesis, inference and simplification of signal transduction networks,
Bioinformatics, Volume 24, Number 2, pp. 293-295, January 2008
 NO
NO