SRM Collider 1.4.2
:: DESCRIPTION
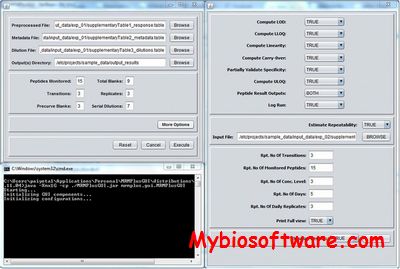
The SRMCollider is a program that will take your input transitions and compare them to all other transitions in a given background proteome and find interferences.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows /MacOsX
- Python
- C++ Compiler
:: MORE INFORMATION
Citation
Mol Cell Proteomics. 2012 Aug;11(8):540-9. doi: 10.1074/mcp.M111.013045. Epub 2012 Apr 24.
A computational tool to detect and avoid redundancy in selected reaction monitoring.
Röst H1, Malmström L, Aebersold R.