IdeogramBrowser 0.20.4
:: DESCRIPTION
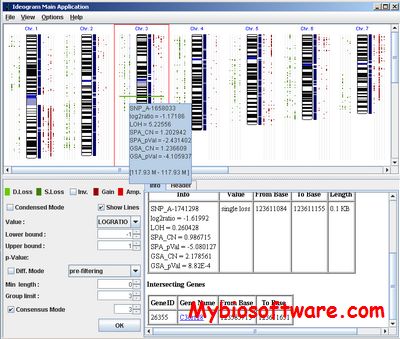
IdeogramBrowser was specifically designed for use with the Affymetrix SNP arrays. It provides an interactive karyotypic visualization of multiple aberration profiles and direct links to GeneCards.
::DEVELOPER
Medical Systems Biology, University of Ulm
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/MacOsX/Linux
- Java
:: MORE INFORMATION
Citation
André Müller, Karlheinz Holzmann and Hans A. Kestler
Visualization of genomic aberrations using Affymetrix SNP arrays
Bioinformatics (2007) 23 (4): 496-497.