PatternLab for proteomics V
:: DESCRIPTION
PatternLab for proteomics is a one-stop shop computational environment for analyzing shotgun proteomic data. Its modules provide means to pinpoint proteins/peptides that are differentially expressed and those that are unique to a state. It can also cluster the ones that share similar expression profiles in time-course experiments, as well as help in interpreting results according to Gene Ontology.
::DEVELOPER
Laboratory for Computational and Strucutural Proteomics – Fiocruz
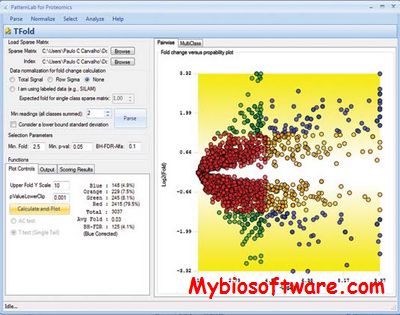
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Paulo C. Carvalho, John R. Yates III, Valmir C. Barbosa
Analyzing Shotgun Proteomic Data with PatternLab for Proteomics
Current Protocols in Bioinformatics Unit 13.13 DOI: 10.1002/0471250953.bi1313s30