BioDownloader 2.2.1.0
:: DESCRIPTION
BioDownloader is a program for downloading and/or updating files from ftp/http servers. The program has unique features that are specifically designed to deal with bioinformatics data files and servers (large file sets, selective file retrieval, built-in download settings repository, wizard for post- download file processing, simultaneous download of multiple files). Besides the intuitive BioDownloader’s features that allow the user to use it out-of-the-box, provides the experienced user a vast set of fine-tunning options and advanced features.
::DEVELOPER
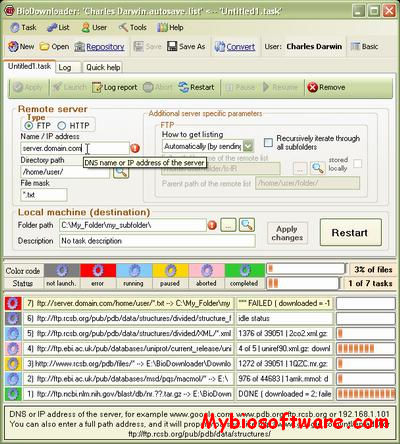
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Maxim V. Shapovalov; Adrian A. Canutescu; Roland L. Dunbrack Jr.
BioDownloader: Bioinformatics downloads and updates in a few clicks.
Bioinformatics 2007; doi: 10.1093/bioinformatics/btm120

 NO
NO